The Analytic Doping Model makes it possible to express doping profiles as a function of the local coordinate system, which can be rotated with respect to the global coordinate system if required. This is suitable to achieve profiles that are convenient to define in relation to the coordinate axes. The dopant distribution can be defined using either the user defined or box methods.
The box method allows a block-shaped region of constant doping to be defined, along with a decay profile away from the region. This is useful for approximating some physical doping techniques, such as diffusion processes, which distribute dopants away from regions of high concentration resulting in characteristic decay profiles. The location of the region is defined by specifying either the corner or center coordinate using the global coordinate system. If a local rotated coordinate system is used within the feature, the orientation of the block rotates around this specified coordinate. A profile is selected from the Dopant profile away from uniform region list. The decay length scale of the profile is controlled by either specifying a control length directly or by specifying a junction depth. The junction depth specifies the distance, from the boundary of the uniformly doped region, where the dopant concentration is equal to the value specified in the
Background doping concentration input. This distance can be specified independently for each axis in the component geometry by selecting
Specify different length scales for each direction.
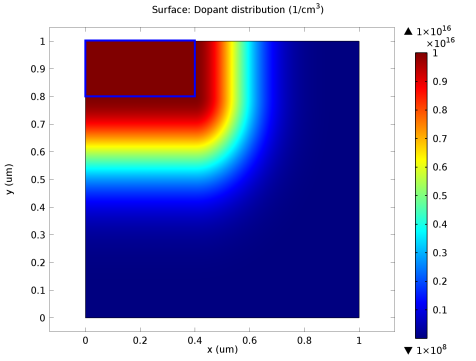
Figure 2-3 is an example of a doping profile created using this method. A rectangular region of uniform doping is specified in the top left of the geometry, and a Gaussian profile is selected away from this region with different junction depths in the
x and
y directions.