
|
|
•
|
mphopen to load the model .mph file.
|
|
•
|
mphmean to evaluate the average concentration in the reactor.
|
|
•
|
mphinterp to evaluate the concentration at specific locations.
|
|
•
|
mphglobal to evaluate global quantities in the model.
|
|
•
|
mphplot to display plots.
|
|
1
|
Start COMSOL with MATLAB.
|
|
•
|
Enter each command, starting at step 2 below, at the MATLAB command line.
|
|
•
|
Paste the full model script, included in the section Model M-File, into a text editor, then save the file with a “.m” extension, and finally run this file in MATLAB.
|
|
5
|
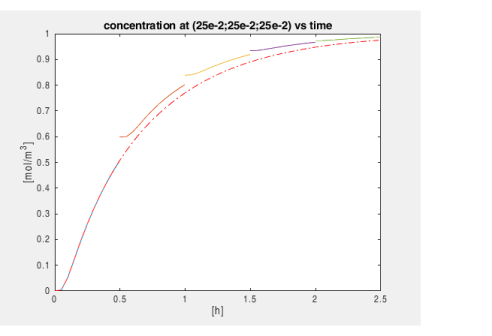
To obtain data for the plot shown in Figure 2 extract the concentration value at the specified location. Use the function mphinterp as shown below:
|
|
7
|
To retrieve the current stop time from the solution, use the command mphglobal according to:
|
|
15
|
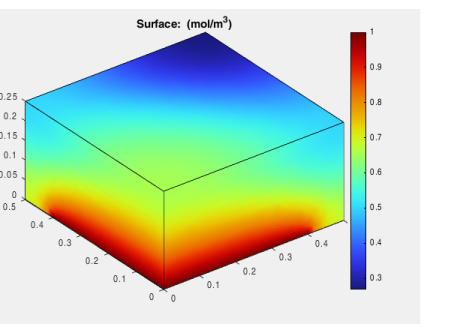
To reproduce the plot in Figure 1, you can also define a plot group in the model:
|
|
1
|
|
2
|
|
3
|
Click Add.
|
|
4
|
Click
|
|
5
|
|
6
|
Click
|
|
1
|
|
2
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
|
1
|
|
2
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
|
6
|
|
1
|
In the Model Builder window, under Component 1 (comp1)>Transport of Diluted Species (tds) click Transport Properties 1.
|
|
2
|
|
3
|
|
1
|
|
3
|
|
4
|
|
1
|
|
3
|
|
4
|
|
5
|
|
1
|
|
2
|
|
3
|
|
5
|
|
6
|
|
1
|
|
2
|
|
3
|
|
1
|
|
2
|
Browse to a directory which path is set in MATLAB and enter domain_activation_llmatlab in the File name text field.
|
|
3
|
Click Save.
|