
|
|
1
|
|
2
|
|
3
|
Click Add.
|
|
4
|
Click
|
|
5
|
|
6
|
Click
|
|
1
|
|
2
|
|
3
|
|
4
|
Browse to the model’s Application Libraries folder and double-click the file nonideal_cstr_parameters.txt.
|
|
1
|
|
2
|
In the Settings window for Reaction Engineering, type Reaction Engineering - CSTR 1 in the Label text field.
|
|
3
|
|
4
|
|
5
|
|
6
|
Locate the Reactor section. Find the Mass balance subsection. From the Volumetric rate list, choose Generic.
|
|
7
|
|
1
|
|
2
|
|
4
|
|
1
|
|
2
|
|
3
|
|
1
|
|
2
|
|
3
|
|
1
|
|
2
|
|
3
|
|
4
|
|
1
|
|
2
|
|
3
|
|
4
|
|
1
|
|
2
|
|
3
|
|
4
|
Locate the Feed Inlet Concentration section. In the Feed inlet concentration table, enter the following settings:
|
|
1
|
|
2
|
|
3
|
|
4
|
Locate the Feed Inlet Concentration section. In the Feed inlet concentration table, enter the following settings:
|
|
5
|
|
1
|
|
2
|
|
3
|
In the Settings window for Reaction Engineering, type Reaction Engineering - CSTR 2 in the Label text field.
|
|
4
|
|
1
|
In the Model Builder window, expand the Component 1 (comp1)>Reaction Engineering - CSTR 2 (re2) node, then click Initial Values 1.
|
|
2
|
|
3
|
|
4
|
|
1
|
|
2
|
|
3
|
|
4
|
Locate the Feed Inlet Concentration section. In the Feed inlet concentration table, enter the following settings:
|
|
1
|
|
1
|
|
2
|
|
3
|
In the Parameter table, enter the following settings:
|
|
4
|
Click
|
|
5
|
In the Parameter table, enter the following settings:
|
|
1
|
|
2
|
|
3
|
Click Browse.
|
|
4
|
Browse to the model’s Application Libraries folder and double-click the file nonideal_cstr_data.csv.
|
|
5
|
Click Import.
|
|
1
|
|
2
|
|
1
|
|
2
|
|
3
|
|
4
|
Locate the Physics and Variables Selection section. Select the Modify model configuration for study step check box.
|
|
5
|
In the Physics and variables selection tree, select Component 1 (comp1)>Reaction Engineering - CSTR 1 (re1)>Parameter Estimation 1.
|
|
6
|
Click
|
|
7
|
|
1
|
|
2
|
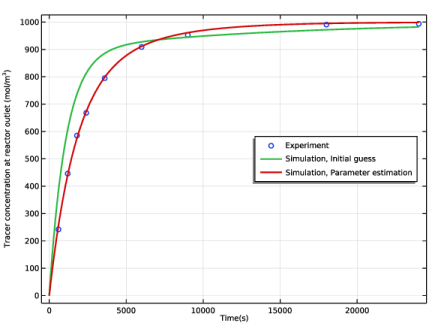
In the Settings window for 1D Plot Group, type Concentration in Real Reactor in the Label text field.
|
|
3
|
|
4
|
|
5
|
In the associated text field, type Time(s).
|
|
6
|
|
7
|
In the associated text field, type Tracer concentration at reactor outlet (mol/m<sup>3</sup>).
|
|
8
|
|
1
|
In the Model Builder window, expand the Concentration in Real Reactor node, then click Experiment 1 Data.
|
|
2
|
|
3
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
|
6
|
|
7
|
|
9
|
|
1
|
|
2
|
|
3
|
|
4
|
In the associated text field, type Tracer concentrations (mol/m<sup>3</sup>).
|
|
5
|
|
1
|
|
2
|
In the Settings window for Global, click Add Expression in the upper-right corner of the y-Axis Data section. From the menu, choose Component 1 (comp1)>Reaction Engineering - CSTR 2>re2.c_T - Concentration - mol/m³.
|
|
3
|
|
4
|
|
6
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
|
1
|
|
2
|
|
3
|
|
4
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
|
6
|
|
1
|
|
2
|
|
1
|
|
2
|
|
3
|
Locate the Data section. From the Dataset list, choose Study 2: Parameter estimation/Solution 2 (sol2).
|
|
4
|
Locate the Legends section. In the table, enter the following settings:
|
|
5
|
|
1
|
|
2
|
|
3
|
|
4
|
|
1
|
|
2
|
|
3
|
|
4
|
|
5
|
Click Replace Expression in the upper-right corner of the Expressions section. From the menu, choose Component 1 (comp1)>Reaction Engineering - CSTR 1>alpha - Global control variable alpha.
|
|
6
|
Click Add Expression in the upper-right corner of the Expressions section. From the menu, choose Component 1 (comp1)>Reaction Engineering - CSTR 1>beta - Global control variable beta.
|
|
7
|
Click
|